Abstract
Molecularly targeted therapeutic and imaging strategies directed at aberrant signaling pathways in pancreatic tumor cells may improve the poor outcome of pancreatic ductal adenocarcinoma (PDA). Therefore, relevant molecular targets need to be identified. Methods: We collected publicly available expression profiles of patient-derived normal pancreatic tissue (n = 77) and PDA samples (n = 103). Functional genomic messenger RNA profiling was applied to predict target upregulation on the protein level. We prioritized these targets based on current status of preclinical therapeutic and imaging evaluation in PDA. Results: We identified 213 significantly upregulated proteins in PDA compared with normal pancreatic tissue. We prioritized mucin-1, mesothelin, γ-glutamyltransferase 5, and cathepsin-E as the most interesting targets, because studies already demonstrated their potential for both therapeutic and imaging strategies in literature. Conclusion: This study can assist clinicians and drug developers in deciding which theranostic targets should be taken for further clinical evaluation in PDA.
- pancreatic ductal adenocarcinoma (PDA)
- pancreatic cancer
- theranostic approach
- targeted molecular therapy
- targeted molecular imaging
- genetic profiling
- biomarker
Pancreatic ductal adenocarcinoma (PDA) is the fourth leading cause of cancer-related mortality worldwide (1). Despite extensive surgery and improved chemotherapeutic regimens, the prognosis of PDA remains poor. Because symptoms often occur late in the disease process, most patients present with locally advanced or even metastatic disease, resulting in a 5-y overall survival rate of only approximately 8% (1). Solely patients with local disease are candidates for curative surgical treatment. Despite the curative intent, the 5-y survival in the surgically treated patients is still as low as 20% (2). This poor survival is partially caused by the rapid development of metastases shortly after surgery. Most likely, this is due to microscopic dissemination that was already present at the time of surgery. Once distant metastases are present, the best available palliative chemotherapy regimen with the best overall survival rate is a combination of fluorouracil, leucovorin, irinotecan, and oxaliplatin. However, the overall survival benefit is modest, and the toxicity is significant (3).
In contrast to the traditional working mechanism of chemotherapy, which has a cytotoxic effect on all rapidly dividing cells, molecularly targeted therapies more selectively target aberrant cell signaling pathways that drive tumor growth. Therefore, in general molecularly targeted therapies are expected to be more tumor specific, which could enhance therapy efficacy and decrease side effects. However, patients who are likely to benefit from a particular targeted therapy have to be selected carefully, and target overexpression needs to be demonstrated. To date, target expression is determined by immunohistochemistry on tissue biopsies, which are prone to be biased by sampling error due to heterogeneity of tumors and metastases. Theranostics, which integrate diagnostics and therapeutics by fluorescent or radioactive labeling of drugs, can provide insight in pharmacokinetics, tumor uptake, and biodistribution of drugs that might be used for clinical decision making and individualized management of disease.
To enable a theranostic approach in PDA patients, there is an unmet need for identification and prioritization of relevant targets. To this end, we used the recently developed method of functional genomic messenger RNA (FGmRNA) profiling to predict overexpression of target antigens on the protein level (4). FGmRNA profiling is capable of correcting a gene expression profile of an individual tumor for physiologic and experimental factors, which are considered not to be relevant for the observed tumor phenotype and characteristics.
The aim of this study was to identify potential target antigens in PDA using FGmRNA profiling that will assist clinicians and drug developers in deciding which theranostic targets should be taken for further evaluation in PDA. Subsequently, an extensive literature search was performed to prioritize these potential target antigens for their use in a theranostic approach in the near future.
MATERIALS AND METHODS
FGmRNA Profiling: Identification of Upregulated Genes in PDA
Data Acquisition
We collected publicly available raw microarray expression data from the Gene Expression Omnibus for the affymetrix HG-U133 plus 2.0 and the HG-U133A platforms (5). We used automatic filtering on relevant key words with subsequent manual curation to include patient-derived PDA samples and normal pancreatic tissue. Cell line samples were deemed irrelevant and excluded for further analysis.
Sample Processing
Noncorrupted raw data files were downloaded from the Gene Expression Omnibus for the selected samples. After removal of duplicate files, preprocessing and aggregation of raw data files were performed with Affymetrix Power Tools (version 1.15.2), using apt-probe set-summarize and applying the robust multiarray average algorithm. Sample quality control was performed using principal component analysis as previously described (6).
FGmRNA Profiling
For a detailed description of FGmRNA-profiling, we refer to Fehrmann et al. (4). In short, we analyzed 77,840 expression profiles of publicly available samples with principal component analysis and found that a limited number of transcriptional components capture the major regulators of the messenger RNA transcriptome. Subsequently, we identified a subset of transcriptional components that described nongenetic regulatory factors. We used these nongenetic transcriptional components as covariates to correct microarray expression data and observed that the residual expression signal (i.e., FGmRNA profile) captures the downstream consequences of genomic alterations on gene expression levels.
Class Comparison
We performed a genome-wide class comparison analysis (Welch’s t test) between FGmRNA profiles of normal pancreatic tissue and PDA to identify genes with upregulated FGmRNA expression, which we considered a proxy for protein expression. To correct for multiple testing, we performed this analysis within a multivariate permutation test (1,000 permutations) with a false-discovery rate of 1% and a confidence level of 99%. This will result in a list of significant upregulated genes, which contains (with a confidence level of 99%) no more than 1% false-positives.
Literature Search on Protein Expression
To compare targets identified with the class comparison with known protein expression in PDA, we performed a literature search. PubMed was searched for articles published in English from conception until February 2017. The following search terms were used: HUGO gene symbol of the target under investigation in combination with ‘pancreatic cancer’, ‘expression’, and ‘immunohistochemistry’. The cellular location and function of the protein product of the gene was explored at http://www.genecards.org.
Target Prioritization for Theranostic Approaches in PDA Based on FGmRNA Profiling
The prioritization process consisted of consulting the drug–gene interaction database (DGIdb) to select targets with a drug–gene interaction, current status of preclinical evaluation of therapeutic drugs directed at the protein, and current status of preclinical evaluation of imaging tracers directed at the protein.
Consulting DGIdb to Identify Drug–Gene Interactions
The DGIdb, accessible at dgibd.genome.wustl.edu, integrates data from 13 resources that include disease-relevant human genes, drugs, drug–gene interactions, and potential druggability (7). Identified targets in the class comparison were explored in the DGIdb to get insight into drug–gene interactions to enable selection of targets for which a drug is available, or targets that are potential according to their membership in gene categories associated with druggability.
Current Status of Therapeutic Efficacy at PubMed and Clinicaltrials.gov
Targets for which a drug–gene interaction was reported by the DGIdb were reviewed in the literature to determine the current status of drugs targeting these genes in clinical translation. We explored the efficacy of drugs targeting the protein in pancreatic cancer; the efficacy of drugs targeting the protein in patients with other cancer types, because these therapies might be relatively easily translated to pancreatic cancer patients; and the knowledge in preclinical studies. PubMed was searched for articles published in English from conception until February 2017, and clinicaltrials.gov was explored for current (ongoing) clinical trials. PubMed was searched using the (1) combination of HUGO gene symbol of the target under investigation; ‘pancreatic AND OR cancer’; and ‘therapy’, or (2) HUGO gene symbol, ‘pancreatic AND OR cancer’.
Current Status of Evaluation of Imaging Targets at PubMed and Clinicaltrials.gov
All targets with a drug–gene interaction were reviewed in literature to prioritize targets that are the furthest in clinical translation and have proved to be a suitable imaging target. An additional PubMed search was executed for articles published in English from conception until February 2017 to determine whether the downstream proteins of these genes are suitable as molecular imaging targets. We used the following search combinations: ‘HUGO gene symbol’; ‘pancreatic AND OR cancer’; and ‘imaging’.
RESULTS
FGmRNA Profiling: Identification of Upregulated Genes in PDA
Supplemental Table 1 (supplemental materials are available at http://jnm.snmjournals.org) shows the datasets that were obtained from the Gene Expression Omnibus. In total, 180 pancreatic samples were identified, which are derived from 16 individual experiments; these samples consisted of 103 PDA and 77 normal pancreatic samples. Class comparison analysis, with multivariate permutation testing (false-discovery rate, 1%; confidence level 99%, 1,000 permutations), resulted in a set of 213 unique genes with significant FGmRNA overexpression in PDA. Supplemental Table 2 contains the class comparison for all genes.
Literature-Based Protein Expression Data for Identified Top 50 Targets Identified with FGmRNA Profiling
On the basis of published immunohistochemistry results of the top 50 upregulated PDA genes as described in Supplemental Table 3, 17 of 50 genes have a known downstream protein overexpression in human PDA samples. The downstream protein overexpression of 5 of 50 genes is described in other solid cancer types and therefore these genes could be of interest for PDA. For 27 of 50 upregulated genes in PDA, no data are available on protein expression in human cancers and therefore might be interesting for preclinical validation in the near future.
Prioritization of Potential Theranostic Targets in PDA
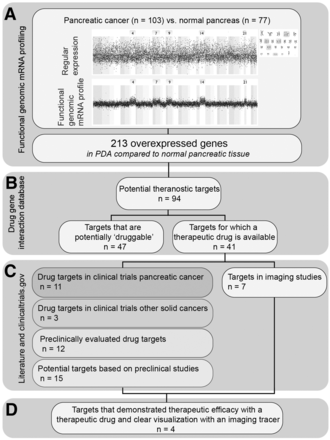
Figure 1 shows the complete prioritization process. Ninety-four of 213 upregulated genes in PDA have a known drug–gene interaction according to DGIdb. Downstream proteins of 41 of 94 genes are currently investigated as a drug target for cancer treatment in clinical trials or in preclinical studies (Fig. 2). Eleven of 41 genes are investigated as antineoplastic drug targets in clinical pancreatic cancer trials, 3 of 41 genes are investigated as antineoplastic drug targets in clinical trials involving other solid cancer types, and 12 of 41 genes are evaluated as antineoplastic drug targets in preclinical in vitro and in vivo cancer models; for 15 of 41 genes, no antineoplastic drugs are currently available that target the downstream proteins, but the literature indicated involvement in cancer development. In addition, downstream proteins of 7 of 41 genes are currently described in the context of molecular imaging. We highlighted the studies evaluating the prioritized targets for molecular imaging purposes in pancreatic cancer or in advanced clinical translation in Supplemental Table 4; a summary of the therapeutic studies can be found in Supplemental Table 5.
Study flowchart shows workflow for identification of current most potential targets for theranostic approaches in future PDA management. (A) We performed FGmRNA profiling to predict protein overexpression in PDA compared with normal pancreatic tissue. Known interaction with antineoplastic drugs was explored at the DGIdB (B), and we explored current status of preclinical evaluation of therapeutic and imaging strategies directed at the antigen (C). (D) We determined most potential theranostic targets on the basis of progress in clinical translation in both imaging and therapy to enable theranostic approaches in PDA on short term.
Potential theranostic target genes based on drug–gene interaction database divided per cellular localization, per evaluation status. (A) Drug targets investigated in clinical trials in PDA patients. (B) Drug targets investigated in clinical trials in other cancer types. (C) Drug targets evaluated in preclinical studies. (D) Potential clinical targets that are currently not evaluated. Italics = targets investigated in vitro; white underline = targets evaluated in imaging studies; bold = most potential theranostic targets.
Thymocyte Differentiation Antigen 1—Rank 1
Molecular ultrasound imaging using microbubbles targeting the membrane protein thymocyte differentiation antigen 1 detected tumors in a transgenic PDA mouse model with a diameter of only several millimeters in size, which were visualized with a 3-fold-higher signal than in normal pancreatic tissue (8).
Cathepsin-E (CTSE)—Rank 8
Ritonavir tetramethyl-BODIPY (RIT-TMB) is an optical imaging agent based on a Food and Drug Administration–approved protease inhibitor. RIT-TMB showed CTSE-specific imaging in a PDA cell line (9). Another CTSE-activatable fluorescence imaging probe demonstrated specific detection of CTSE activity in a PDA mouse model, in which the fluorescence signal in the tumor was 3-fold higher than in background tissue (10).
γ-Glutamyltransferase 5 (GGT5)—Rank 10
The cell membrane–bound enzyme GGT5 can be targeted by the optical imaging probe γGlu-HMRG, which is only fluorescent after cleavage by GGT5 (11). γGlu-HMRG was topically applied on surgical breast cancer specimens to assess the surgical margin. Tumors even smaller than 1 mm could be discriminated from normal mammary gland tissue (12). In mouse models for colon cancer and disseminated peritoneal ovarian cancer, tumors could be clearly visualized 1 min after topical administration (11,13).
Mucin-1 (MUC1)—Rank 41
The downstream cell membrane protein of MUC1 is reported to be overexpressed in 96% of the PDA cases. The 111In-labeled monoclonal antibody PAM4 targeting MUC1 is suitable for single-photon emission tomography. In a clinical phase I trial, 111In-PAM4 showed specific uptake of pancreatic cancer lesions (14). More recently, the MUC1-specific optical imaging tracer Ab-FL-Cy5.5, which is a dual-labeled MUC1-targeting antibody conjugated to both a far-red dye and a green dye, demonstrated specific uptake and in vivo visualization of ovarian cancer xenografts (15). The MUC1 aptamer–based tracer APT-PEG-MPA showed that tracer uptake in the tumor correlated well with MUC1 expression levels in MUC1-overexpressing hepatocellular carcinoma and lung carcinoma cells in a xenograft mouse model (16). Furthermore, the anti-MUC1 optical imaging tracer CT2 demonstrated selective targeting of pancreatic cancer in vitro, and in a pancreatic cancer orthotopic xenograft model tumors smaller than 5 mm could be detected (17).
Mesothelin (MSLN)—Rank 110
The overexpression of the cell membrane protein MSLN has been described in up to 86%–100% of PDA cases (18–20). In a clinical phase I imaging trial, the 89Zr-labeled MSLN antibody 89Zr-MMOT0530A was administered in 11 metastatic cancer patients, 7 with PDA and 4 with ovarian cancer. In all patients, at least 1 tumor lesion could be visualized (21). In addition to this PET tracer, a MSLN-specific tracer has been developed for single-photon emission tomography. 111In-labeled amatuximab was investigated in 6 patients, 2 of whom had PDA. In all patients, at least 1 tumor lesion could be discriminated from its reference background (22).
DISCUSSION
In this study, we were able to use FGmRNA profiling on a substantial set of normal pancreatic tissue and PDA tissue to predict protein overexpression for a large set of targets and identified 213 upregulated targets in PDA, containing 41 currently druggable targets with the potential for a theranostic approach in PDA patients.
The selection of suitable targets for imaging or therapy is complex. The ideal target is highly overexpressed at the cell membrane of tumor cells and has a limited expression at the cell membrane of normal cells. Immunohistochemistry is a widely used method for the determination of protein expression at a cellular level. However, it is time consuming and it demands many resources including access to formalin-fixed and paraffin-embedded tissue samples of interest. Moreover, differences in execution of the staining protocol and scoring methods make it difficult to compare immunohistochemistry results from different studies. In contrast, FGmRNA profiling enabled us to efficiently analyze and directly compare many genes as the predicted overexpression is determined for each gene with the same methodology including a large set of normal pancreatic tissue samples as a reference to determine the threshold for overexpression. Therefore, FGmRNA profiling has the advantage over immunohistochemistry for the first selection of new therapeutic and imaging targets. FGmRNA profiling previously demonstrated it can guide clinicians and researches in selecting targets that need further preclinical validation, enabling a more efficient use of limited resources (20,23).
Theranostic drugs might be used for clinical decision making by enabling visualization of molecular characteristics of the tumor to stratify patients for the most optimal targeted therapy. Besides, theranostics can aid in monitoring treatment effects, helping clinicians to adjust therapy dose or to switch to another targeted drug. On the basis of the current status of preclinical evaluation of therapeutic drugs and imaging tracers directed at downstream proteins of genes identified with FGmRNA profiling, we prioritized MUC1, MLSN, GGT5, and CTSE as the current most potential theranostic targets. These targets have already shown great potential to serve as a target for both therapy and imaging in the literature, indicating that these drugs have already made progress in the clinical translation process and are potentials for clinical translation in pancreatic cancer patients on the short term. Other targets (e.g., thymocyte differentiation antigen 1) first need to be validated as suitable targets; either therapeutic drugs or imaging tracers need to be designed and subsequently investigated in preclinical studies before theranostic agents targeting these proteins can be investigated in clinical trials.
Beside theranostic targets, FGmRNA profiling can guide researchers and clinicians in selecting targets for molecular imaging probes. After prioritization, only 7 of the 41 currently druggable targets are described in the context of molecular imaging, indicating the great potential of our results for development of favorable molecular imaging probes. In PDA, molecular imaging might enhance disease staging by enabling visualization of small PDA lesions, possibly leading to optimized selection of patients who will benefit from curative surgery. Clinical trials already demonstrated the feasibility of molecular fluorescence imaging in identifying micrometastases in peritoneal metastasized ovarian and colon cancer patients by targeting the folate α-receptor and vascular endothelial growth factor A (24–26). Besides, molecular imaging can be used to better assess the extent of the primary tumor during PDA surgery and evaluate essential resection planes. In PDA patients, 2 clinical trials are currently registered that evaluate intraoperative molecular fluorescence imaging: targeting vascular endothelial growth factor A (NCT02743975) and the epidermal growth factor receptor (NCT02736578). FGmRNA profiling predicted no overexpression of these proteins, which might negatively influence the likelihood of success compared with targets highly rated by FGmRNA profiling. However, in addition to alteration in gene expression levels, mutation occurring in genes can result in different activation or functionality of the gene. This phenomenon is not captured by FGmRNA profiling, but could be relevant for certain tumor phenotypes observed in PDA. For newly identified targets that are not highly rated in the FGmRNA profiling, we advise solid validation in ex vivo models and preclinical models to confirm the validity of the target.
Furthermore, by fluorescent or radioactive labeling of therapeutic drugs, molecular imaging can provide insight in pharmacokinetics, tumor uptake, and biodistribution, which harbors the potential for drug development to select probes with great therapeutic potential and to support optimal dosing and determine uptake in critical organs to anticipate toxicity. This is especially relevant in PDA because a desmoplastic reaction surrounding the tumor increases interstitial fluid pressure impairing drug delivery. Therefore, molecular imaging might help to determine which probes might be successfully translated into theranostic agents.
CONCLUSION
This study provides a data-driven prioritization and overview of imaging and therapeutic targets. The presented data can assist clinicians, researchers, and drug developers in deciding which therapeutic or imaging targets should be taken for further clinical evaluation in PDA. This might help to improve disease outcome of PDA patients in the short term.
DISCLOSURE
This work was financially supported by grants from the Dutch Cancer Society/Alpe d'HuZes (RUG 2013-5960 to Rudolf S. N. Fehrmann and RUG 2012-5416 to Wouter B. Nagengast), the Dutch Organization for Scientific Research (NWO-VENI grant 916-16025), and a Mandema Stipendium to Rudolf S. N. Fehrmann and Wouter B. Nagengast. Marjory Koller and Gooitzen M. van Dam report grants from the FP-7 Framework Programme BetaCure (grant no. 602812) during the study. No other potential conflict of interest relevant to this article was reported.
Footnotes
Published online Oct. 19, 2017.
- © 2017 by the Society of Nuclear Medicine and Molecular Imaging.
REFERENCES
- Received for publication July 20, 2017.
- Accepted for publication October 12, 2017.